Published in Computational Methods in Systems Biology, September 16–18, 2024.
https://link.springer.com/chapter/10.1007/978-3-031-71671-3_11
Abstract
Alzheimer’s disease is expected to reach a prevalence of 152 million people worldwide caused by the aggregation of amyloid β-proteins leading to apoptosis of neurons and loss of cognitive function. Although there is no effective treatment for this disease, molecules such as scyllo-inositol have been shown to be promising. Bacillus subtilis has been proposed as a suitable organism for the production of scyllo-inositol.
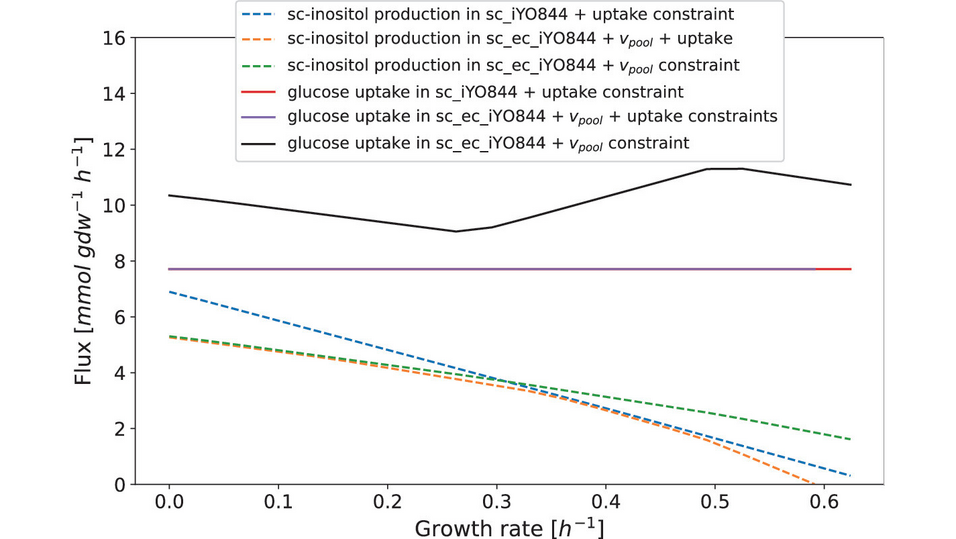
Metabolic computational models have proven useful in the prediction of the production of a metabolite. However, most genome-scale metabolic models lack detailed parameters and tend to overestimate the production of a metabolite with respect to the consumption of medium resources. In order to reduce the solution space and, hence, obtain a more realistic model, additional constraints from experimental data can be added to the model.
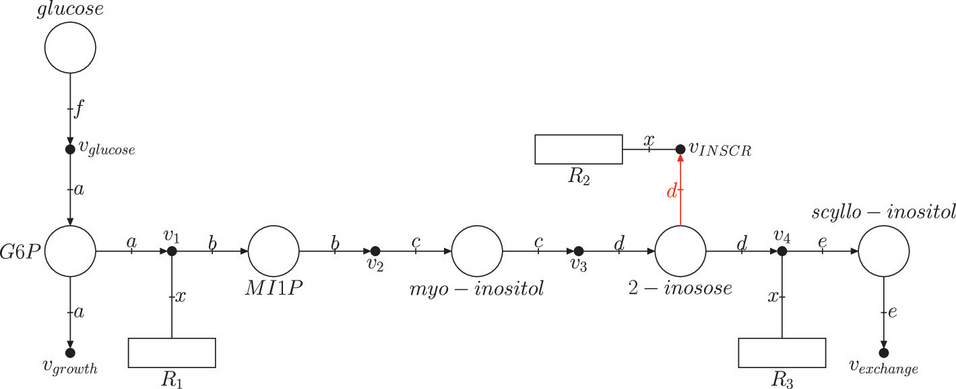
This work exploits the modeling capabilities of Flexible Nets to model the production of scyllo-inositol in a genome-scale metabolic model of Bacillus subtilis that has been previously enriched with proteomic and enzymatic data. We assess how these constraints limit the scyllo-inositol production to more realistic levels. Moreover, the integration of different types of constraints allowed us to uncover which one of them limits the production of scyllo-inositol for a given growth rate.