Published in Computational and Structural Biotechnology Journal, Volume 27, 1498 - 1510.
https://doi.org/10.1016/j.csbj.2025.03.040
Highlights
- Flexible Nets combined intracellular fluxes with bioreactor dynamics for modeling.
- The model was validated, capturing key constraints like medium and uptake rates.
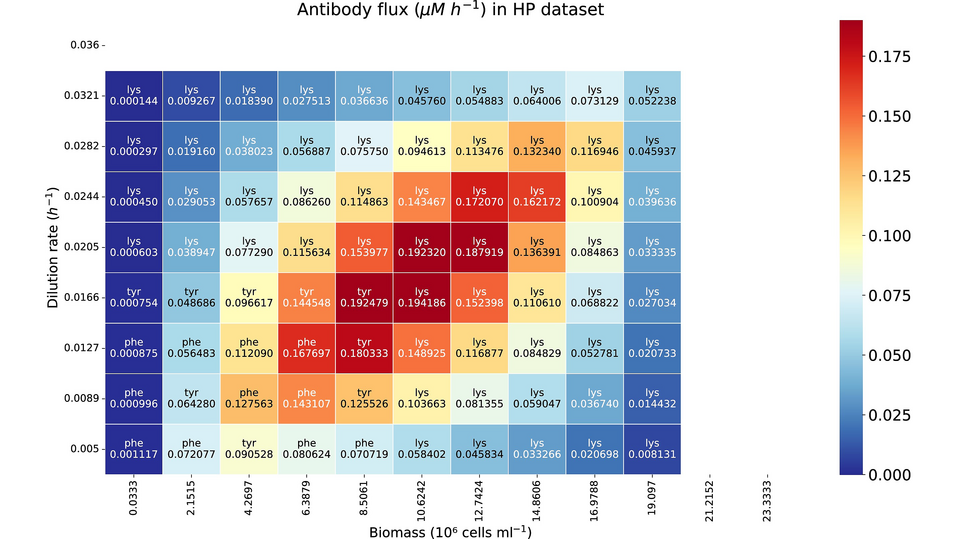
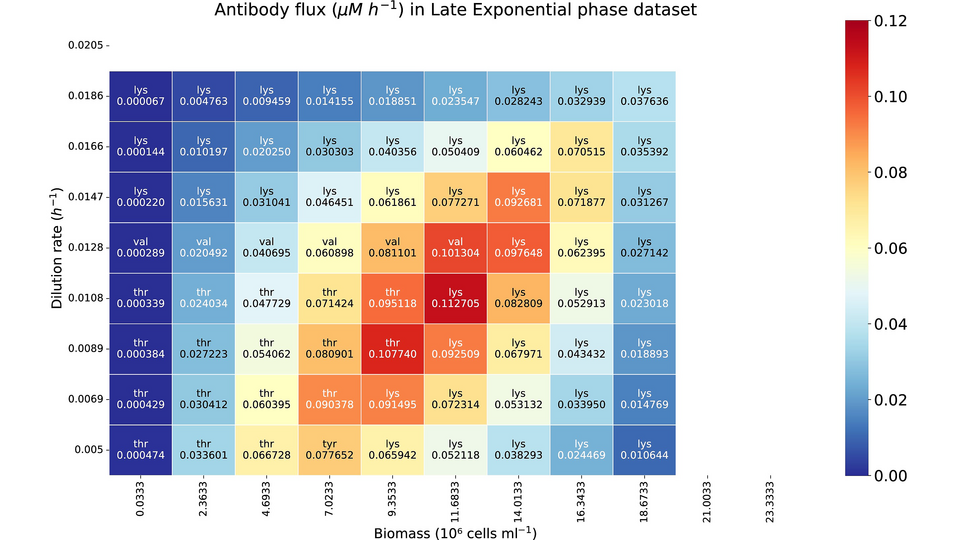
- A trade-off between cell growth and IgG production was detected.
- The model showed improved IgG yields by adjusting the biomass and dilution rate.
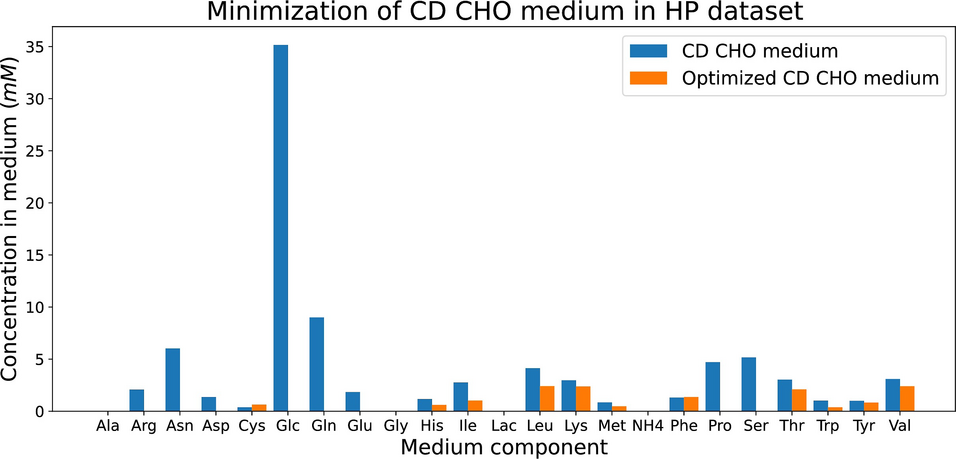
- Including uptake rates provided a framework for IgG production and medium enhancing.
Abstract
Antibodies are therapeutic proteins with many applications in medicine, such as treating viral infections, different types of cancer, and common diseases such as psoriasis and multiple sclerosis. Chinese Hamster Ovary (CHO) cells are the most widely used cells for antibody production due to their well-established use and favorable features. However, the current design of antibody production systems often relies on a “trial and error” approach to manipulate CHO cells. This approach is time-consuming and costly, and can lead to suboptimal process performance. The use of mathematical models has the potential to greatly accelerate and improve the design and optimization of antibody production. Starting from a systematic and formal approach, the aim is to achieve an automatic design of the whole process that allows optimal productivity to be reached. To this end, we develop mathematical models and methods for the design and optimization of antibody manufacturing systems. The mathematical models are based on Flexible Nets (FNs), a modeling formalism that accommodates uncertain parameters and nonlinear dynamics. FNs enable the development of comprehensive models that encompass both the metabolic network of CHO cells and the dynamics of the bioreactor in which the cells are cultured. Thus, by integrating macroscopic variables (e.g. dilution rate, substrate concentration, cell density, etc.) with microscopic variables (such as intracellular metabolic fluxes), our model represents a multi-scale system and facilitates global optimization.