Published in Briefings in Bioinformatics, Volume 26, Issue 2, March 2025, bbaf188.
https://doi.org/10.1093/bib/bbaf188
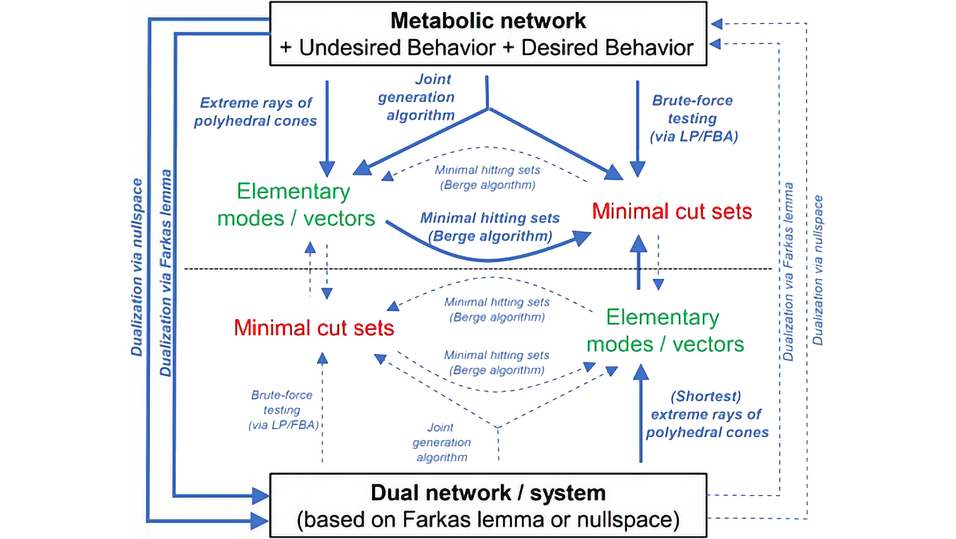
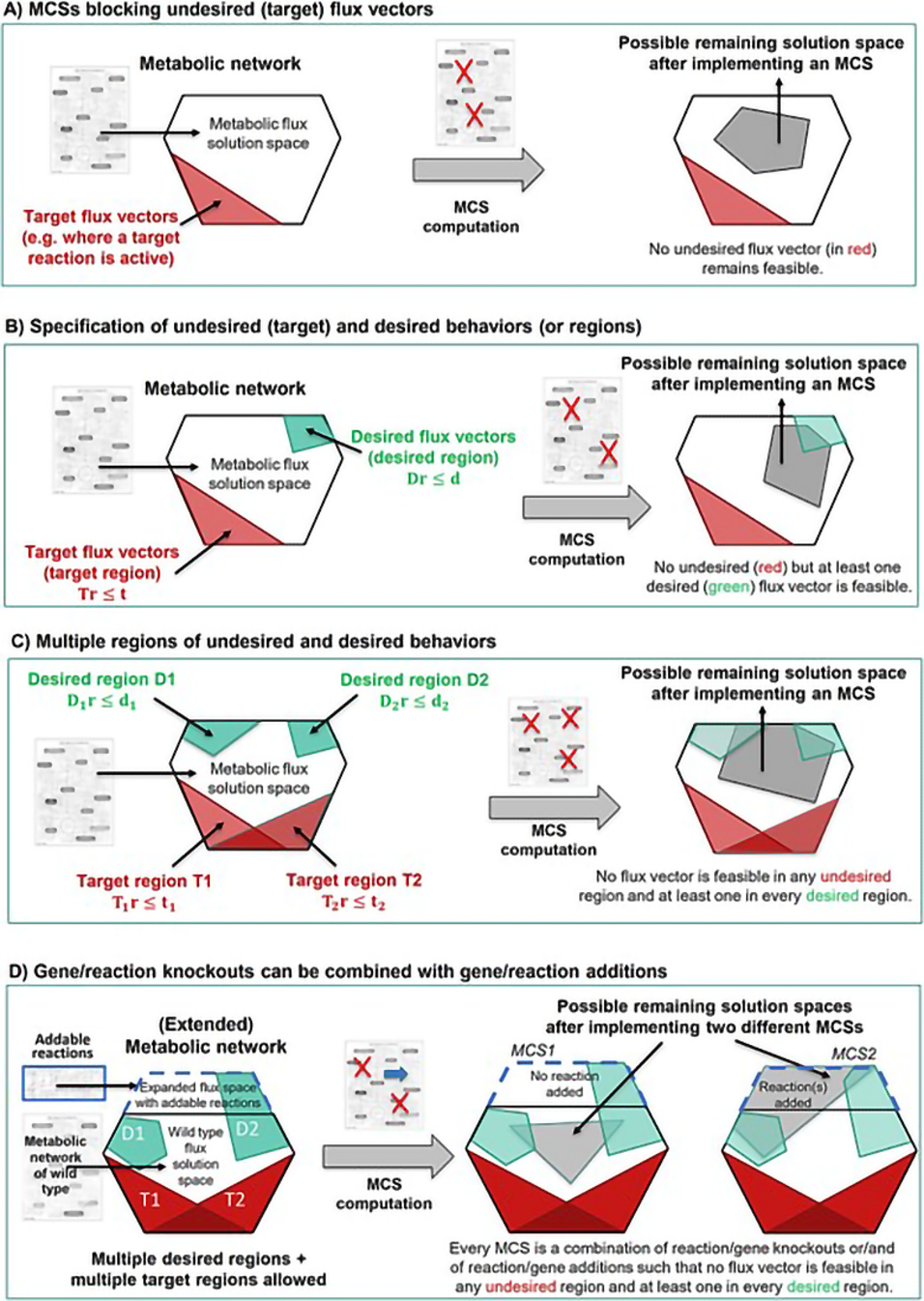
Minimal cut sets (MCSs) have emerged as an important branch of constraint-based metabolic modeling, offering a versatile framework for analyzing and engineering metabolic networks.
Over the past two decades, MCSs have evolved from a theoretical concept into a powerful tool for identifying tailored metabolic intervention strategies and studying robustness and failure modes of metabolic networks. Successful (experimental) applications range from designing highly efficient microbial cell factories to targeting cancer cell metabolism.
This review highlights key conceptual and algorithmic advancements that have transformed MCSs into a flexible methodology applicable to metabolic models of any size. It also provides a comprehensive overview of their applications and concludes with a perspective on future research directions.
The review aims to equip both newcomers and experts with the knowledge needed to effectively leverage MCSs for metabolic network analysis and design, therapeutic targeting, and beyond.