Published in Bioinformatics, 40, 4, April 2024, btae153.
doi.org/10.1093/bioinformatics/btae153
See the code on GitHub.
Abstract
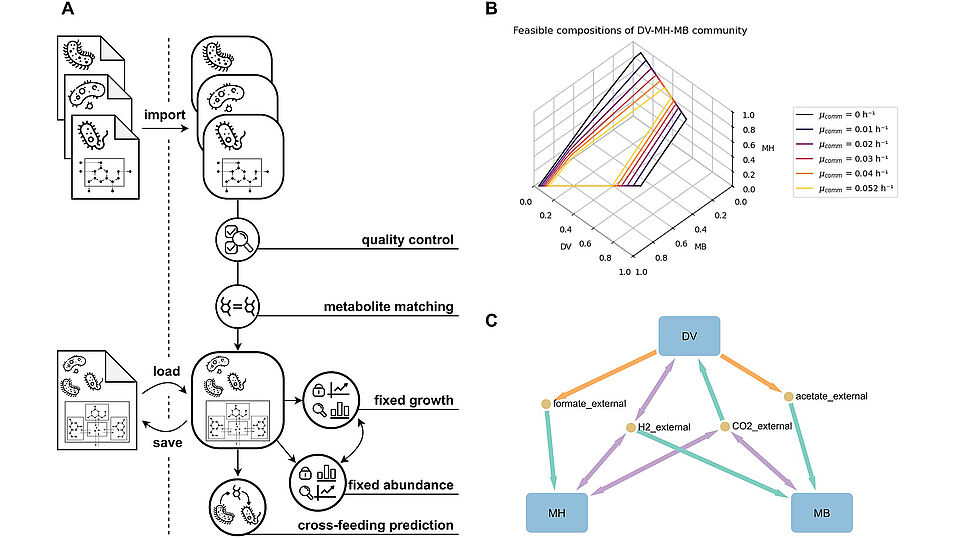
PyCoMo is a python package for quick and easy generation of genome-scale compartmentalized community metabolic models that are compliant with current openCOBRA file formats. The resulting models can be used to predict (i) the maximum growth rate at a given abundance profile, (ii) the feasible community compositions at a given growth rate, and (iii) all exchange metabolites and cross-feeding interactions in a community metabolic model independent of the abundance profile; we demonstrate PyCoMo’s capability by analysing methane production in a previously published simplified biogas community metabolic model.
Availability and implementation
PyCoMo is freely available under an MIT licence at http://github.com/univieCUBE/PyCoMo, the Python Package Index, and Zenodo.
Further explanations may be found under https://github.com/mpredl.